Network SIR
Graph Sampling and Disease Spread In Small Population Networks
My team and I explored a dataset and publication regarding small-population networks. The dataset contained both
face-to-face contact data (two individuals directly interacting) and co-presence contact data (two individuals in the
same vicinity). We attempted to first assess whether or not co-presence data can be used as a proxy for face-to-face
data. Co-presence, though cheaper and easier to gather, presents a much more dense network than face-to-face networks.
We implemented a number of graph sub-sampling algorithms to show that co-presence data still encodes the face-to-face
graph structure, and can thus be successfully used as a proxy. We decided upon the Metropolis-Hastings sampling
algorithm as it provided a non-naive sampling approach at relatively fast compute, while maintaining graph structure.
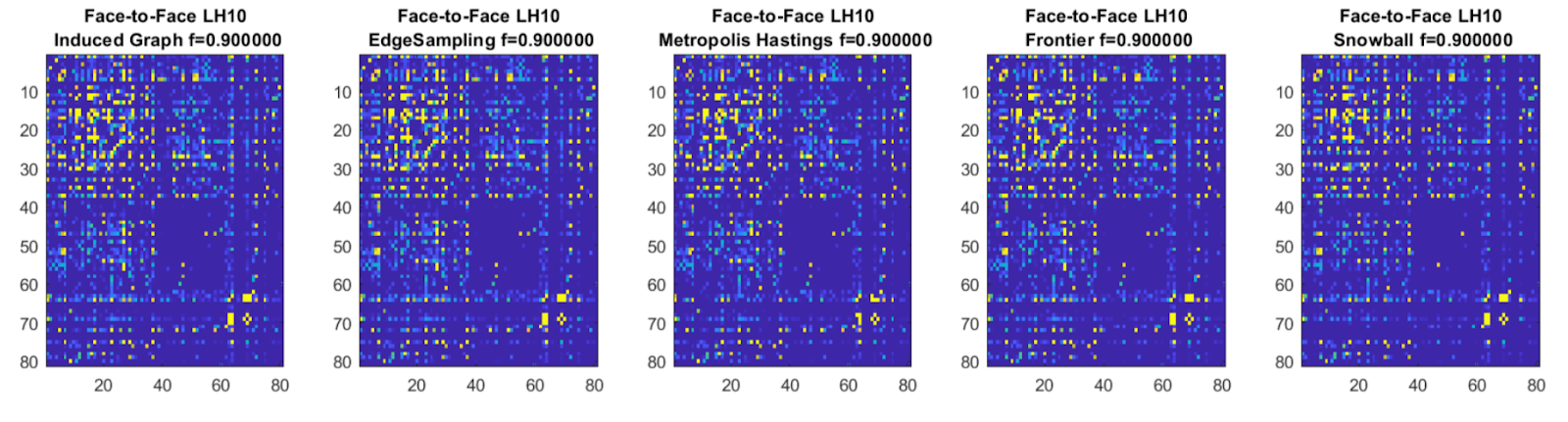
F2F Graph Sampling |
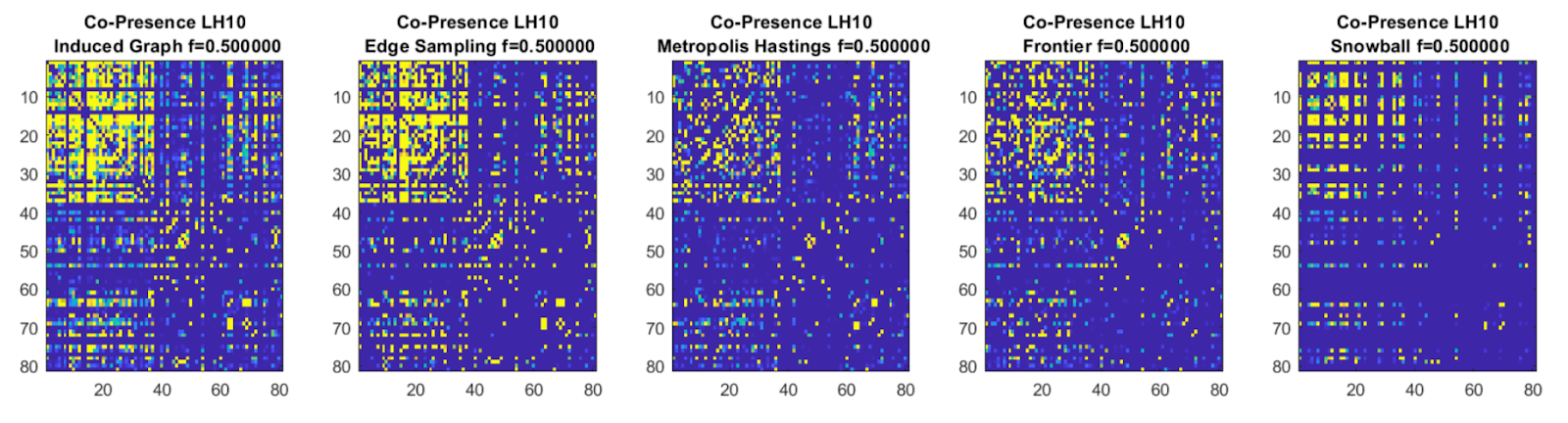
Co-Presence Graph Sampling |
The below imgs show the comparison of graph size metrics (density, volume, etc.) for F2f and Co-presence networks. The image on the right shows that once the graphs are subsampled, the co-presence network holds similar network topology to that of the face-to-face network.
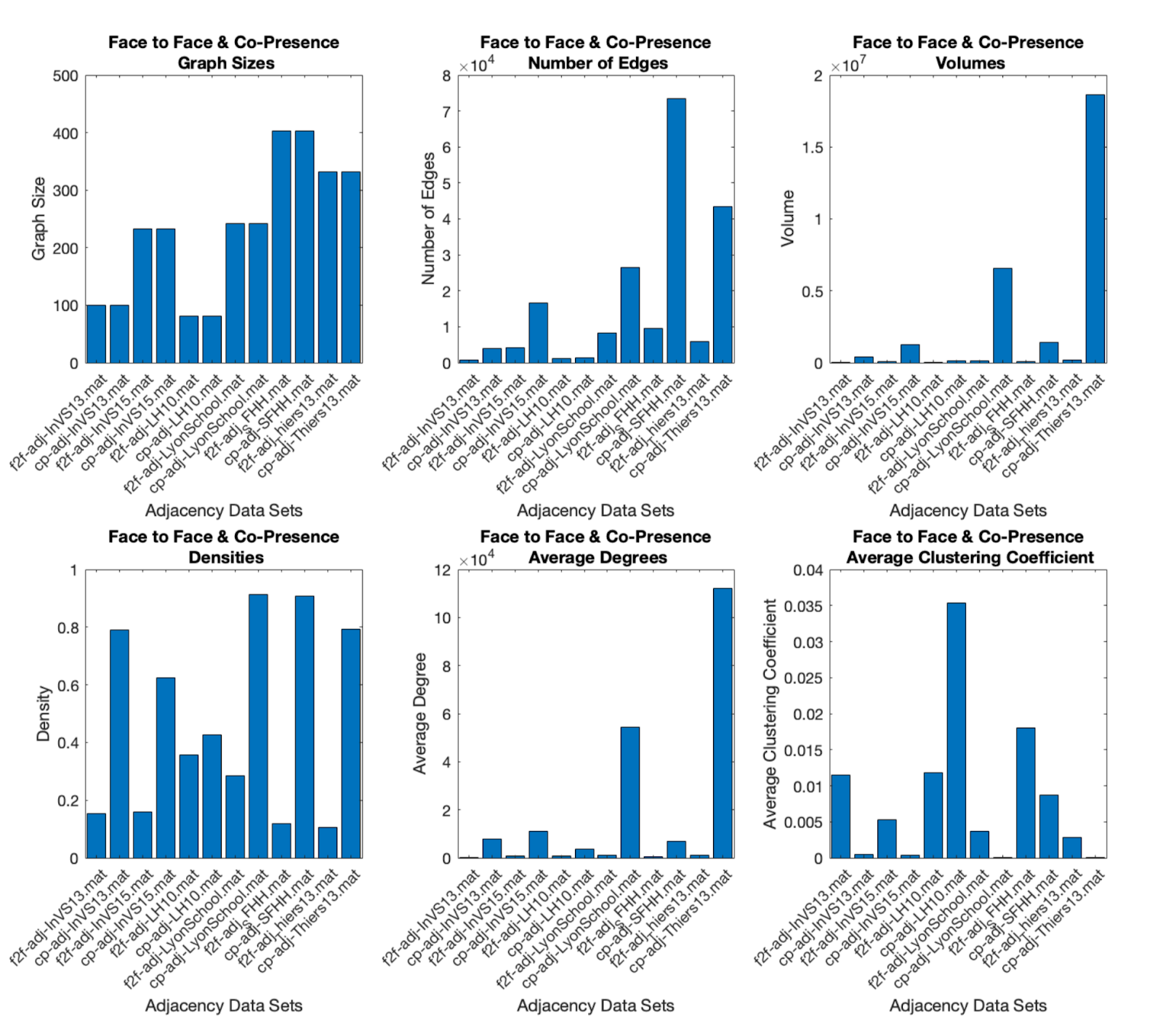
F2F vs Co-P Network Size |
F2F vs Sampled Co-P Graph Structure |
We then apply these subsampled graphs to SIR (Susceptible, Infectious, Recovered) disease spread model and explore a number of vaccination schemes. The baseline SIR model, with no vaccination, can be seen below. We further observe that co-presence, given the data, can represent f2f activity in an SIR with fair accuracy.
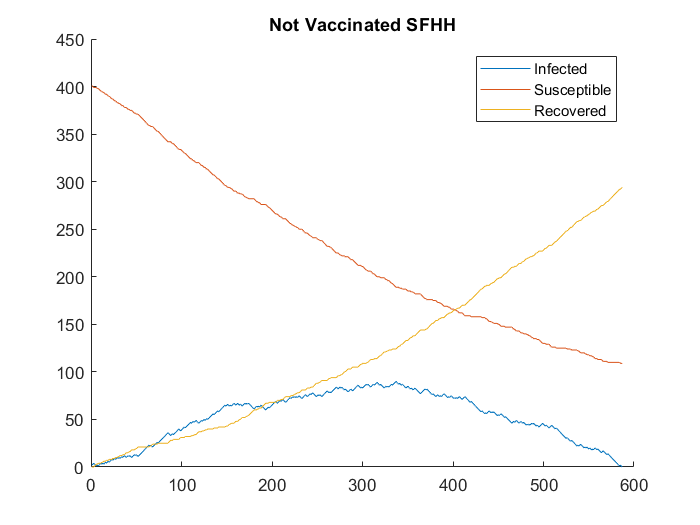
F2F vs Co-P Network Size |
|
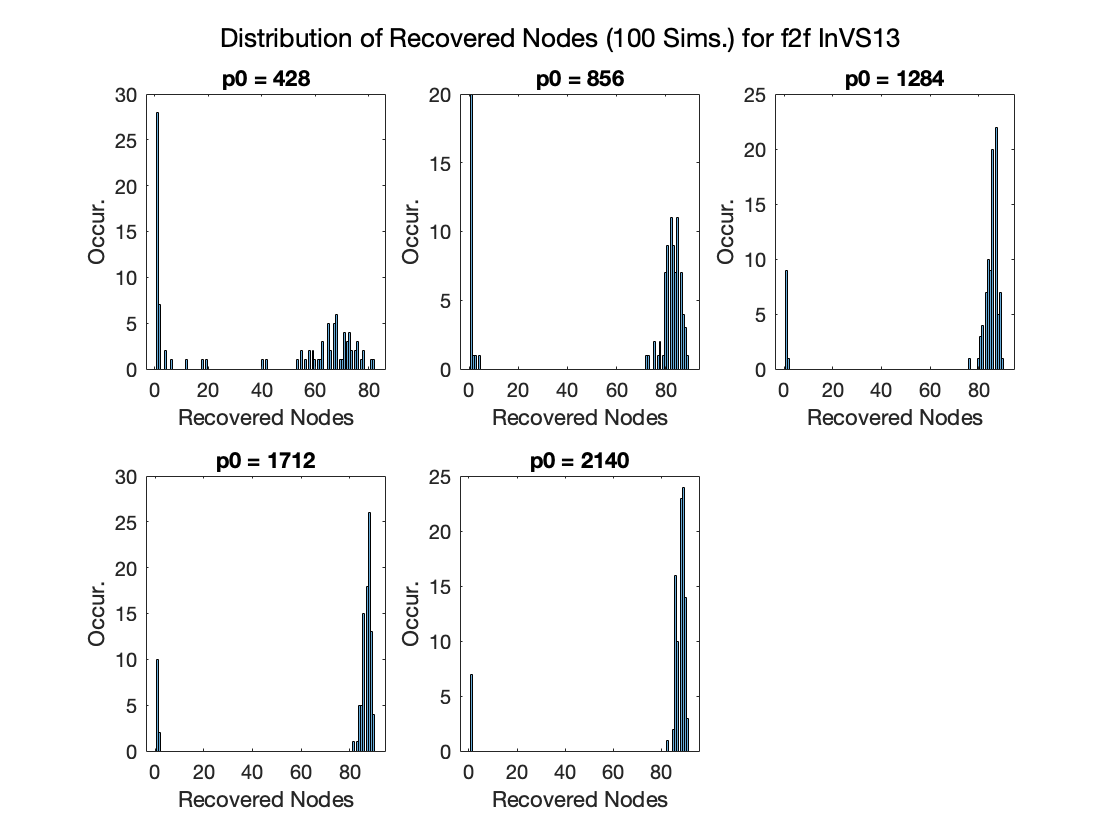
Recovered nodes in F2F Data |

Recovered nodes in Co-P Data |
We finally analyze vaccination in both F2F and Co-Presence networks. We iterate over no vaccination, random vaccination, and vaccination of highly-connected nodes. We report for both F2F and Co-Presence data, that vaccinating nodes with the most connections is the most beneficial scheme. This, of course is a scenario in which highly-connected individuals are known and there is also not enough vaccines for the entire graph-population.
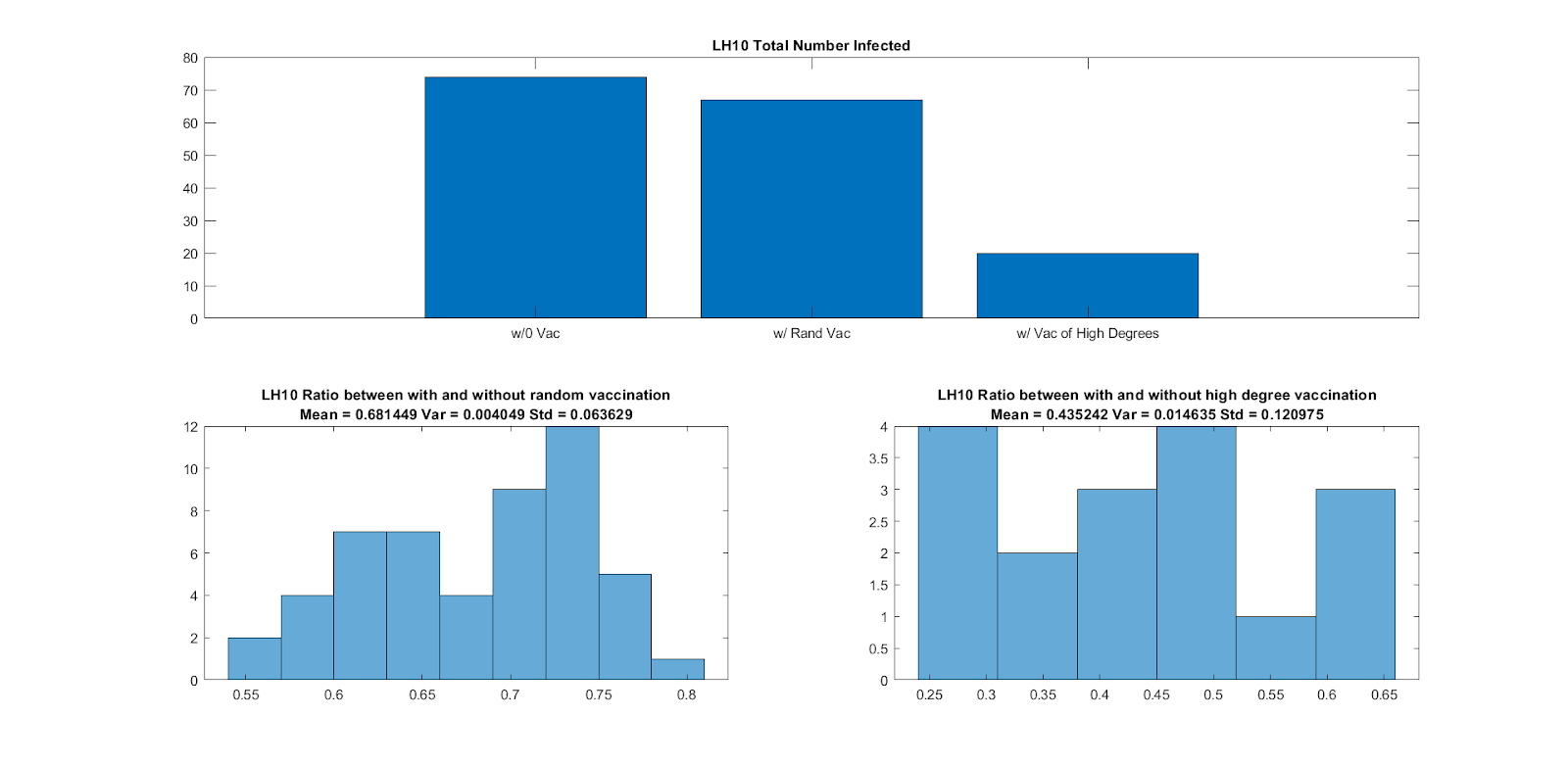
Vaccination in F2F Networks |
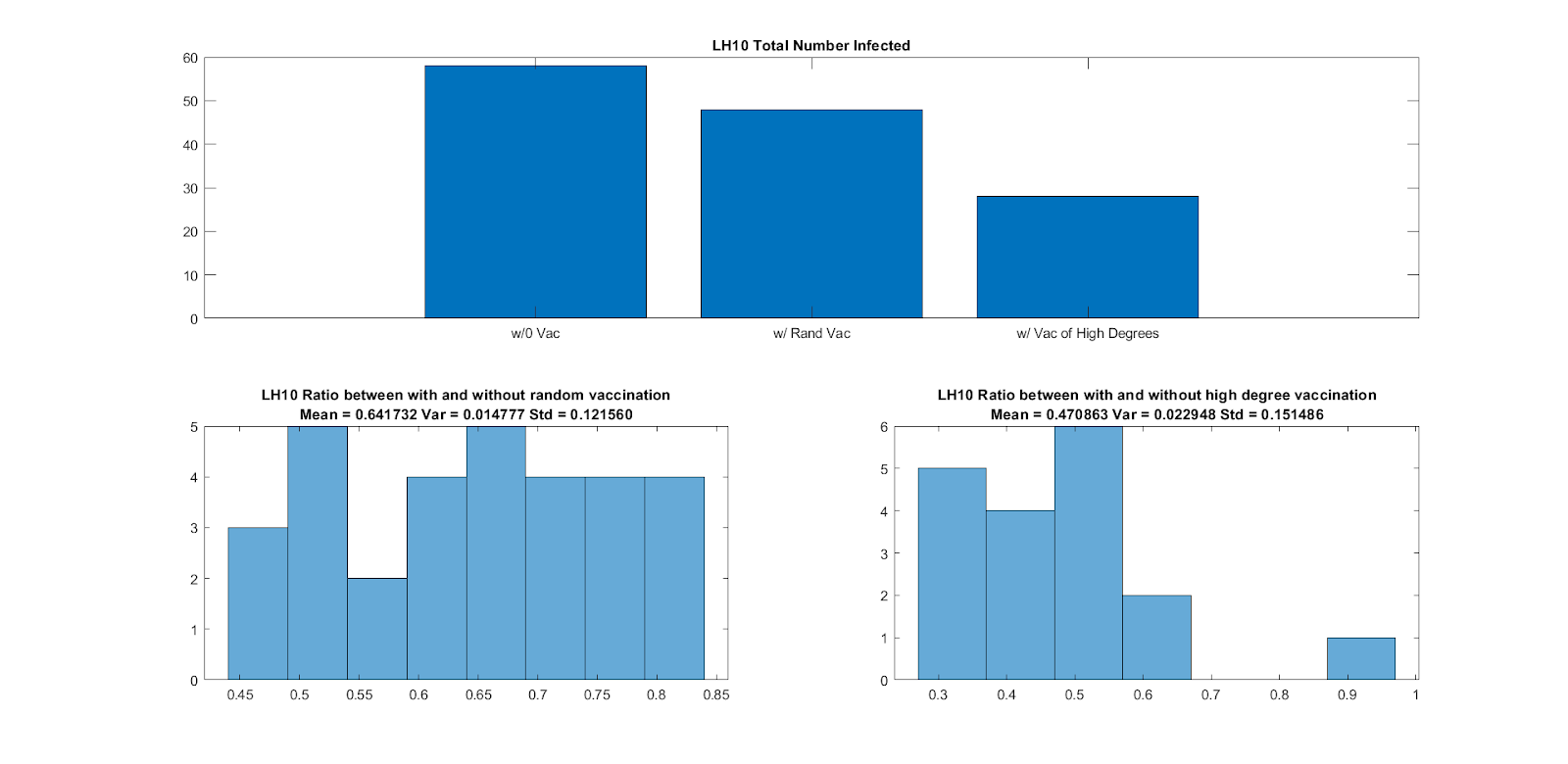
Vaccination in Co-P Networks |
This work mirrors some of the early work used for analyzing the COVID-19 pandemic, and provides a base-level insight for how information and disease propagates through a graph. The vaccination analysis further corroborates known findings regarding disease prevention. The SIR model, and similar models have been thoroughly studied recently with the COVID-19 pandemic.
Additional Material
Collaborators
Karros Huang, Clint Olsen, Francois Meyer